Projects
Dynamic myocardial blood volume mapping with MR Multitasking
This project focuses on developing a fully motion-resolved framework for quantifying myocardial fractional blood volume (fMBV) using MR Multitasking in conjunction with ferumoxytol, an intravascular contrast agent. The Multitasking acquisition produces a highly undersampled dataset that is reconstructed through a low-rank subspace model, yielding separate dimensions for cardiac motion, respiratory motion, and contrast-driven T1 dynamics. Within this latent space, motion-resolved T1 maps are generated across the entire cardiac cycle and across multiple ferumoxytol doses. Subsequently, a compartmental model is fitted to the multi-dose T1 evolution to estimate fMBV and associated water-exchange parameters.
Central to this work are the stabilization of cross-dose and cross-phase alignment of data within the subspace, and the construction of a pipeline capable of producing dynamic, physiologically interpretable fMBV maps without reliance on ECG gating or breath-holding. This framework aims to improve the characterization of myocardial microvascular function and provide a more robust approach to contrast-enhanced quantitative cardiac MRI.

Contrast-free multi-quantitative MRI with MR Fingerprinting
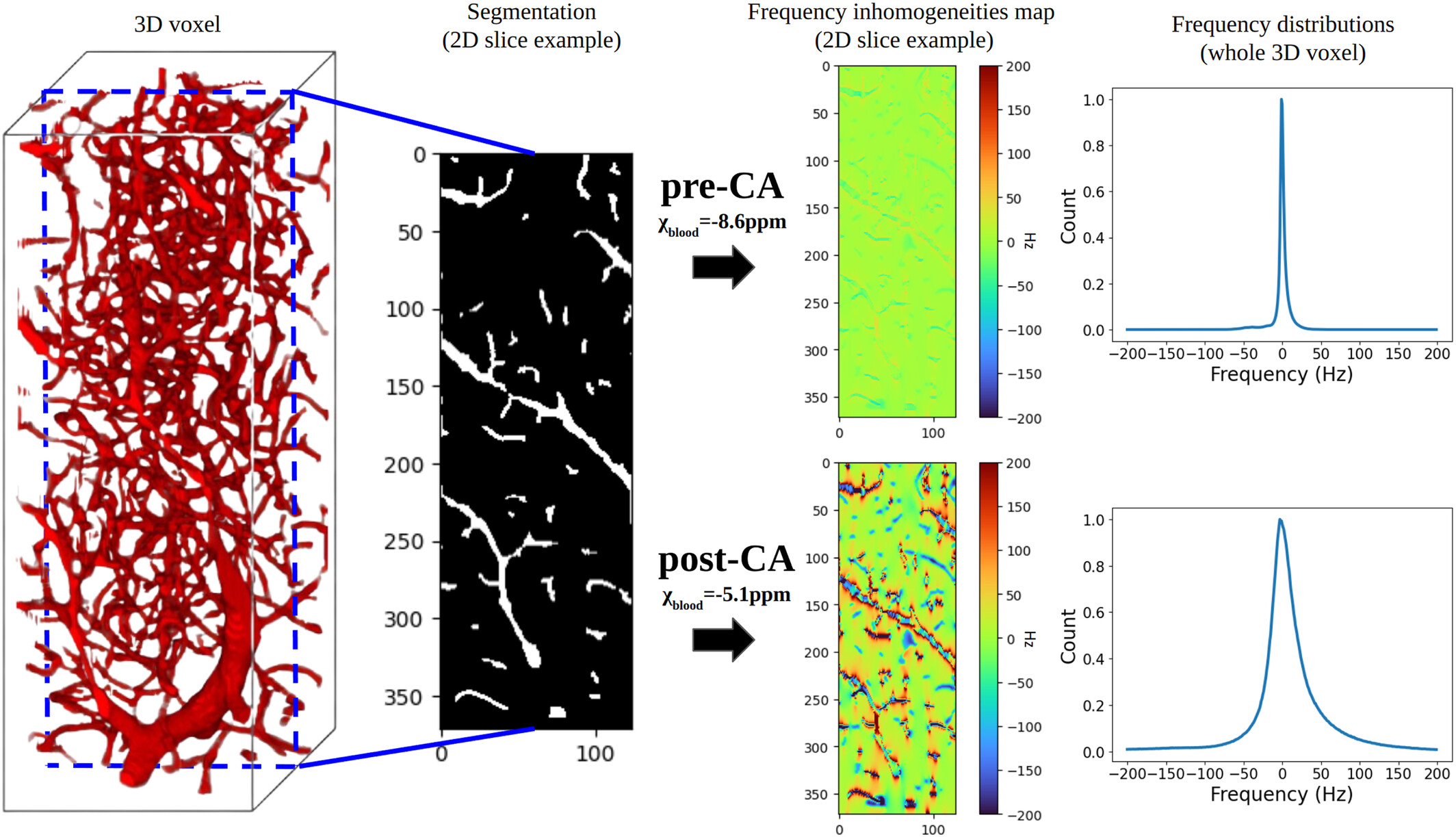
The concept of MRF proposes to reconstruct images by directly comparing in vivo acquisitions and millions of digital simulations that mimic brain tissue (dictionary). This approach is flexible enough to allow access to several MRI parameters simultaneously and to quantify them. As shown in clinical and preclinical studies, when the simulations are based on the physics of NMR (magnetic field, magnetic susceptibility, diffusion, relaxations, etc.) and on biophysics (cellular dimensions and vascular, flow ...), the technique allows the extraction of information on tissue microstructures. These data are essential for the management of patients with acute stroke. Until now, the digital representation of tissues has been based on simple shapes (cells = spheres, blood vessels = cylinders) and the simulation step is time-consuming. In addition, the choice of the 4D sequence is currently based on rough nuclear magnetization trajectories and the comparison step is extremely computer expansive and also time-consuming. Finally, the actual measurements of perfusion and oxygenation parameters are constrained by contrast agent injection.
This project is centered on the development of a contrast-free MR Fingerprinting (MRF) framework for rapid, multidimensional quantitative brain imaging in the context of cerebrovascular diseases. The method relies on matching measured MRF signal evolutions to a comprehensive dictionary of simulations that incorporate detailed NMR physics—including relaxation as well as realistic microvascular and cellular architectural features. By generating high-fidelity digital phantoms of brain tissue and accelerating large-scale reconstruction through advanced dictionary compression or physic-informed neural network, the project enabled the extraction of relaxation, perfusion, and oxygenation biomarkers without the need for exogenous contrast agents. The resulting framework provides quantitative parameters with direct links to tissue microstructure and hemodynamics, offering a non-invasive alternative for assessing neurovascular conditions such as acute stroke.
This framework targets quantitative biomarkers for acute stroke and other neurovascular diseases by linking the MRF signal evolution to underlying tissue microstructure and hemodynamics.
MR-WAVES: MR simulations from 3D realistic microvascular networks in a few seconds, using recurrent neural network and histogram-based approximation
MR-WAVES is a computational framework designed to replace traditional Monte-Carlo spin-dynamics simulations with a recurrent neural network model capable of predicting MR signal evolutions in realistic three-dimensional microvascular geometries. Conventional simulations, while accurate, are computationally intensive and limit the exploration of diverse sequence designs or microvascular configurations. MR-WAVES leverages simulated datasets to train an RNN that learns the temporal evolution of magnetization under varying gradient waveforms, structural geometries, and biophysical conditions. Once trained, the model provides rapid and highly scalable signal predictions, enabling efficient sequence optimization and facilitating large-scale studies on how microvascular architecture influences measurable MRI contrast. This framework offers a practical and interpretable surrogate for complex microvascular physics, bridging detailed biophysical modeling with modern machine-learning-based signal prediction.